Computational tools fuel reconstruction of new and improved bird family tree
An international team of scientists has built the largest and most detailed bird family tree to date—an intricate chart delineating 93 million years of evolutionary relationships between 363 bird species, representing 92% of all bird families. Credit: Jon Fjeldså (drawings) and Josefin Stiller An international team of scientists has built the largest and most detailed […]
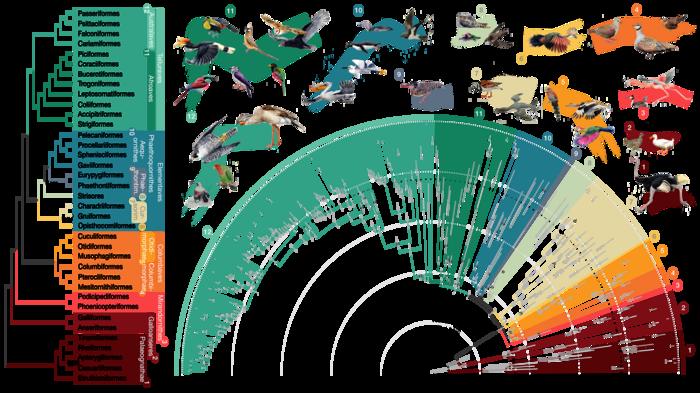
An international team of scientists has built the largest and most detailed bird family tree to date—an intricate chart delineating 93 million years of evolutionary relationships between 363 bird species, representing 92% of all bird families.

Credit: Jon Fjeldså (drawings) and Josefin Stiller
An international team of scientists has built the largest and most detailed bird family tree to date—an intricate chart delineating 93 million years of evolutionary relationships between 363 bird species, representing 92% of all bird families.
The advance was made possible in large part thanks to cutting-edge computational methods developed by engineers at the University of California San Diego, combined with the university’s state-of-the-art supercomputing resources at the San Diego Supercomputer Center. These technologies have enabled researchers to analyze vast amounts of genomic data with high accuracy and speed, laying the groundwork for the construction of the most comprehensive bird family tree ever assembled.
The advance is detailed in two complementary papers published on April 1 in Nature and the Proceedings of the National Academy of Sciences (PNAS). The updated family tree, reported in Nature, revealed patterns in the evolutionary history of birds following the cataclysmic mass extinction event that wiped out the dinosaurs 66 million years ago. Researchers observed sharp increases in effective population size, substitution rates and relative brain size in early birds, shedding new light on the adaptive mechanisms that drove avian diversification in the aftermath of this pivotal event. In the companion paper published in PNAS, researchers closely examined one of the branches of the new family tree and found that flamingos and doves are more distantly related than previous genome-wide analyses had shown.
The work is part of the Bird 10,000 Genomes (B10K) Project, a multi-institutional effort led by University of Copenhagen, Zhejiang University and UC San Diego that aims to generate draft genome sequences for about 10,500 extant bird species.
“Our goal is to reconstruct the entire evolutionary history of all birds,” said Siavash Mirarab, professor of electrical and computer engineering at the UC San Diego Jacobs School of Engineering, who is a co-senior author on the Nature paper, as well as first and co-corresponding author on the PNAS paper.
Piecing together the past
At the heart of these studies lies a suite of algorithms known as ASTRAL, which Mirarab’s lab developed to infer evolutionary relationships with unprecedented scalability, accuracy and speed. By harnessing the power of these algorithms, the team integrated genomic data from over 60,000 genomic regions, providing a robust statistical foundation for their analyses. The researchers then examined the evolutionary history of individual segments across the genome. From there, they pieced together a mosaic of gene trees, which were then compiled into a comprehensive species tree. This meticulous approach enabled the researchers to construct a new and improved bird family tree that delineates complex branching events with remarkable precision and detail, even in cases of historical uncertainty.
“We found that our method of adding tens of thousands of genes to our analysis was actually necessary to resolve evolutionary relationships between bird species,” said Mirarab. “You really need all that genomic data to recover what happened in this certain period of time 65-67 million years ago with high confidence.”
The team’s ability to conduct these analyses on massive datasets was made possible because Mirarab’s lab designed their computational methods to run on powerful GPU machines. They ran their calculations on the Expanse supercomputer at the San Diego Supercomputer at UC San Diego.
“We were fortunate to have access to such a high-end supercomputer,” said Mirarab. “Without Expanse, we would not have been able to run and rerun our analyses on such large datasets in a reasonable amount of time.”
The researchers also looked at the effects of different genome sampling methods on the accuracy of the tree. They showed that two strategies—sequencing many genes from each species, as well as sequencing many species—combined together are important for reconstructing this evolutionary history.
“Because we used a mixture of both strategies, we could test which approach has stronger impacts on phylogenetic reconstruction,” said Josefin Stiller, professor of biology at the University of Copenhagen and lead author of the Nature paper. “We found that it was more important to sample many genetic sequences from each organism than it was to sample from a broader range of species, although the latter method helped us to date when different groups evolved.”
Correcting the past
With the aid of their advanced computational methods, the researchers were also able to shed light on something unusual that they had discovered in one of their previous studies: a particular section of one chromosome in the bird genome had remained unchanged for millions of years, void of the expected patterns of genetic recombination.
This anomaly initially led the researchers to incorrectly group flamingos and doves together as evolutionary cousins, for they appeared closely related based on this unchanged section of DNA. That’s because their previous analysis was based on the genomes of 48 bird species. But by repeating their analysis using the genomes of 363 species, a more accurate family tree emerged that moved doves further from flamingos. Moreover, using six high-quality genomes provided by the Vertebrate Genome Project (VGP)—led by co-author Erich Jarvis, a professor of neurobiology at Rockefeller University—Mirarab and colleagues were able to detect and putatively explain this surprising pattern.
“What’s surprising is that this period of suppressed recombination could mislead the analysis,” said Edward Braun, professor of biology at the University of Florida and co-corresponding author of the PNAS paper. “And because it could mislead the analysis, it was actually detectable more than 60 million years in the future. That’s the cool part.”
Next steps
The impact of this work extends far beyond studying the evolutionary history of birds. The computational methods pioneered by Mirarab’s lab have become one of the standard tools for reconstructing evolutionary trees for a variety of other animals.
Moving forward, the team is continuing their efforts to construct a complete picture of bird evolution. Biologists are working on sequencing the genomes of additional bird species in the hopes of expanding the family tree to include thousands of bird genera. Meanwhile, computational scientists led by Mirarab are refining their algorithms to accommodate even larger datasets to ensure that analyses in future studies are conducted with high speed and accuracy.
Nature paper: “Complexity of avian evolution revealed by family-level genomes.”
PNAS paper: “A region of suppressed recombination misleads neoavian phylogenomics.”
Journal
Nature
DOI
10.1038/s41586-024-07323-1
Article Title
Complexity of avian evolution revealed by family-level genomes
Article Publication Date
1-Apr-2024
What's Your Reaction?