Researchers use machine-learning modeling tools to improve zinc-finger nuclease editing technology
Genome editing is making inroads into biomedical research and medicine. By employing biomolecule modeling tools, a Japanese research team is accelerating the pace and cutting the cost of zinc finger nuclease (ZFN) technology, a primary gene editing tool. Credit: News organizations may use or redistribute this image, with proper attribution, as part of news coverage […]
Genome editing is making inroads into biomedical research and medicine. By employing biomolecule modeling tools, a Japanese research team is accelerating the pace and cutting the cost of zinc finger nuclease (ZFN) technology, a primary gene editing tool.

Credit: News organizations may use or redistribute this image, with proper attribution, as part of news coverage of this paper only.
Genome editing is making inroads into biomedical research and medicine. By employing biomolecule modeling tools, a Japanese research team is accelerating the pace and cutting the cost of zinc finger nuclease (ZFN) technology, a primary gene editing tool.
In a recently published study, researchers from Hiroshima University and the Japanese National Institute of Advanced Industrial Science and Technology demonstrated how machine learning-driven modular assembly systems can improve gene editing.
The study was published on April 10 in the journal Advanced Science.
“Genome editing is a promising tool for the treatment of genetic disorders in a number of different fields,” said Shota Katayama, associate professor in the Genome Editing Innovation Center at Hiroshima University. “By improving the efficiency gene editing technologies, we can achieve greater precision in modifications to the genetic information in living cells.”
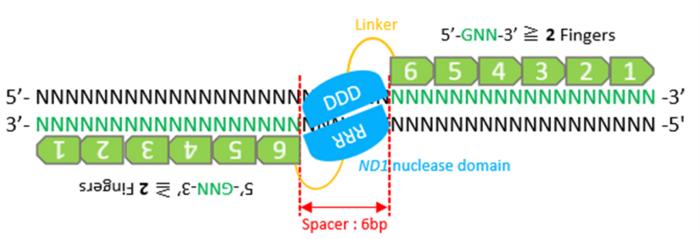
Alongside CRISPR/Cas9 and TALEN, zinc finger nuclease is an important tool in the field of genome editing. Engineered to break certain bonds within the polynucleotide chain of a DNA molecule, these chimeric proteins are made up of two domains fused together: DNA-binding and DNA-cleavage domains. The zinc finger (ZF) protein binding domain recognizes the targeted DNA sequence within the complete genome, while the cleavage domain involves a special DNA-cutting enzymes called ND1 endonucleases.
ZFN present a few advantages over CRISPR/Cas9 and TALEN: first, unlike for CRISPR-Cas9, the patents for ZFNs have already expired, precluding high patent royalties for industrial applications. Secondly, ZFN are smaller, allowing for ZFN-encoding DNA to be easily packaged into a viral vector with limited cargo space for in vivo and clinical applications.
To cut DNA, two ZFNs must be bonded. Therefore, they must be designed in pairs to be functional at any new site. However, constructing functional ZFNs and improving their genome editing efficiency has proved challenging.
“We’ve made huge strides in methods for deriving zinc-finger sets for new genomic targets, but there is still room to improve our approaches to design and selection,” Katayama said.
Selection-based methods can be used to construct assembled ZF proteins, but these methods are labor intensive and time-consuming. An alternative approach for constructing assembled ZF proteins is the assembly of ZF modules using standard molecular biology techniques. This method provides researchers with a much easier method to construct assembled ZF proteins.
However, modularly assembled ZFNs have a small number of functional ZFN pairs with a 94 percent failure rate for the ZFN pairs tested.
In their study, the researchers from Hiroshima University and the Japanese National Institute of Advanced Industrial Science and Technology aimed to create a more efficient, easily constructable zinc finger nuclease for gene editing using publicly available resources in a modular assembly system.
An important consideration in the design of ZFNs is the number of zinc fingers that are required for efficient and specific cleavage. The team hypothesized that the modular assembly of the ZF modules would be useful for constructing ZFNs with five or six fingers.
In their publication, the research team presented a method to increase the efficiency of construction of functional ZFNs and the improvement of their genome editing efficiency using three biomolecule modeling tools: AlphaFold, Coot and Rosetta.
Of the ten ZFNs tested, the researchers obtained two functional pairs. Furthermore, the engineering of ZFNs using AlphaFold, Coot and Rosetta increased the efficiency of genome editing by 5%, demonstrating the effectiveness of engineering ZFNs based on structural modeling.
The research is supported by the Center of Innovation (COI) Program.
Other contributors include Masahiro Watanabe and Yoshio Kato from the National Institute of Advanced Industrial Science and Technology, and Wataru Nomura and Takashi Yamamoto from Hiroshima University.
###
About Hiroshima University
Since its foundation in 1949, Hiroshima University has striven to become one of the most prominent and comprehensive universities in Japan for the promotion and development of scholarship and education. Consisting of 12 schools for undergraduate level and 4 graduate schools, ranging from natural sciences to humanities and social sciences, the university has grown into one of the most distinguished comprehensive research universities in Japan. English website: https://www.hiroshima-u.ac.jp/en
Journal
Advanced Science
DOI
10.1002/advs.202310255
Article Title
Engineering of Zinc Finger Nucleases Through Structural Modeling Improves Genome Editing Efficiency in Cells
Article Publication Date
10-Apr-2024
What's Your Reaction?

































